
Tidybayes and ggdist 3.0 are now on CRAN. There are a number of big changes, including some slightly backwards-incompatible changes, hence the major version bump.
Major changes include:
- Support for slabs with true gradients with varying
alphaorfillin R 4.1. - Improved support for discrete distributions.
- Support for the new posterior package, including the rvar (random variable) datatype.
side,justification, andscaleare now aesthetics instead of parameters, allowing them to vary across slabs within the same geom.- Re-organization of the tidybayes prediction functions, including the
deprecation of
tidybayes::add_fitted_draws()in favor ofadd_epred_draws()andadd_linpred_draws(), and improved support for model types beyond brms and rstanarm. - Slabs are now laid out in a way that makes it easier to line up
geom_slabinterval()with other geoms when usingposition = "dodge". - Various improvements designed to make it easier for other package developers to use ggdist internally.
Support for true gradients in R 4.1
We’ll use the following libraries for the demos below:
library(ggdist)
library(tidybayes)
library(ggplot2)
library(dplyr)
library(distributional)
library(modelr)
library(brms)
theme_set(theme_ggdist())If you are using R >= 4.1, fills and alphas that vary within a slab in
geom_slabinterval() can now be drawn as true gradients rather than segmented
polygons by setting fill_type = "gradient". This substantially improves the
appearance of gradient fills in graphics engines that support it:
dist_df = data.frame(
group = c("a","b","c","d"),
mean = 1:4,
sd = c(1.6,1.4,1.2,1)
)
dist_df %>%
ggplot(aes(x = group, dist = dist_normal(mean, sd), fill = group)) +
stat_dist_gradientinterval(fill_type = "gradient") +
scale_fill_brewer(palette = "Dark2") +
ggtitle(
'stat_dist_gradientinterval(fill_type = "gradient")',
'aes(dist = dist_normal(mean, sd))'
)
Because not all graphics engines support fill_type = "gradient", it is not turned
on by default at the moment. You can omit fill_type = "gradient" and the gradients
will still be displayed, but they make look “choppy”. For more on support for
“true” gradients in R 4.1, see Paul Murrell’s blog post on the topic.
Improved support for discrete distributions
There are various improvements to support for discrete distributions, including better
automatic binwidth selection for discrete distributions in geom_dotsinterval()
and the automatic detection of discrete distributions in stat_dist_slabinterval().
The latter allows discrete analytical distributions to be automatically
displayed as histograms:
tibble(
group = c("a","b","c","d","e"),
lambda = c(13,7,4,3,2)
) %>%
ggplot(aes(x = group)) +
stat_dist_slab(aes(dist = dist_poisson(lambda), fill = stat(pdf))) +
geom_line(aes(y = lambda, group = NA), size = 1) +
geom_point(aes(y = lambda), size = 2.5) +
labs(fill = "Pr(y)") +
ggtitle(
"stat_dist_slab()",
"aes(dist = dist_poisson(lambda), fill = stat(pdf))"
)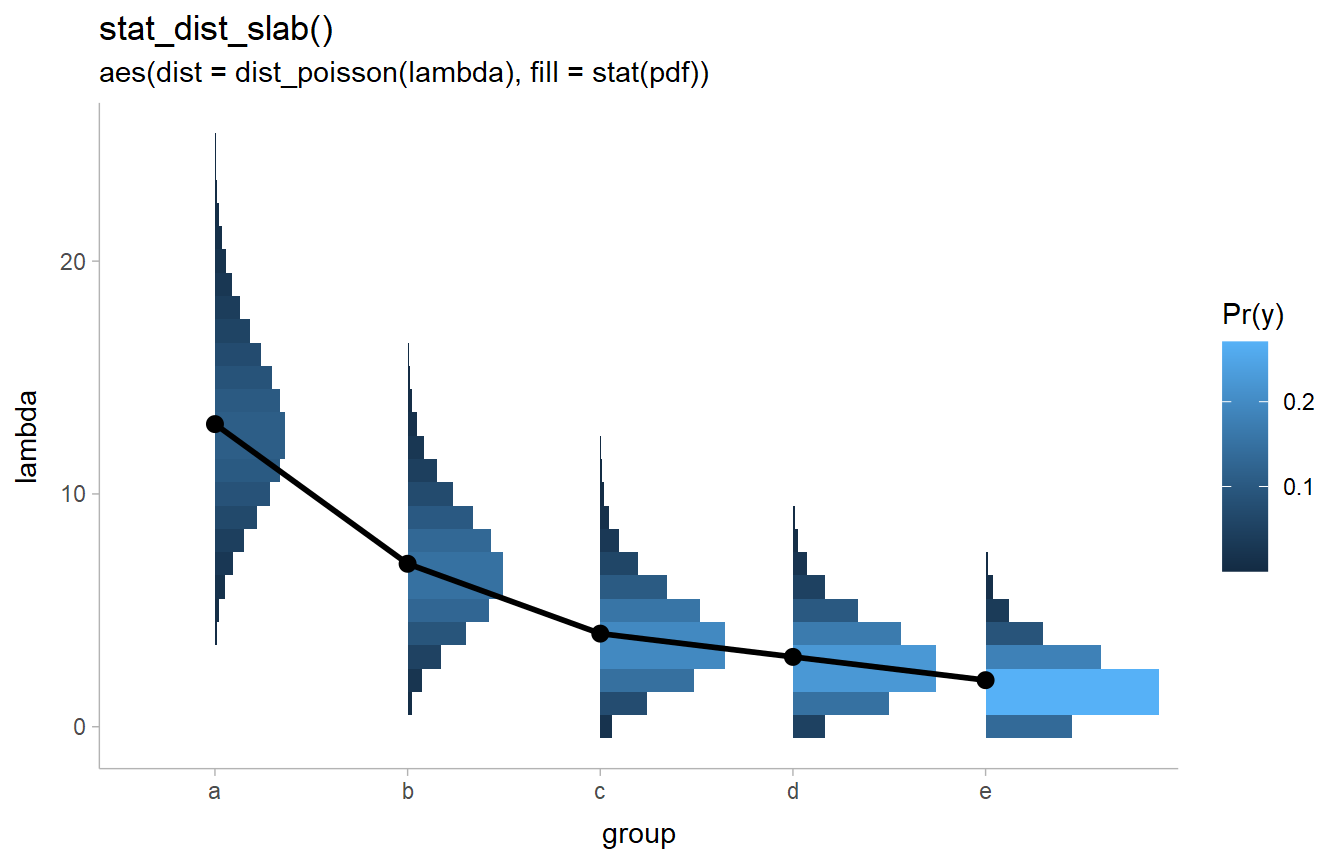
The improved support for discrete distributions in stat_dist_slabinterval()
was precipitated by a discussion
with Isabella Ghement.
Support for the new posterior package
Tidybayes and ggdist now support the posterior
package, and in particular the new rvar
datatype. All of the existing stat_dist... geoms in ggdist can be passed
rvars using the dist aesthetic (just like with objects from
distributional).
Tidybayes also now has a variety of functions suffixed with _rvars instead of
_draws which work similarly to their _draws counterparts. To demo them, let’s
use the palmerpenguins dataset. Here is the body mass of three penguin species
broken down by sex:
data(penguins, package = "palmerpenguins")
penguins %>%
na.omit() %>%
ggplot(aes(y = species, x = body_mass_g, color = sex)) +
stat_dots(position = "dodge", shape = 19, size = 0) +
scale_color_brewer(palette = "Set2", guide = guide_legend(reverse = TRUE))
Males tend to be heavier than females. Let’s fit a simple model to predict sex based on body mass and species:
m_penguin = brm(
# for this simple example I'm not using a varying slope, e.g.
# (body_mass_g|species), though that probably would be better
sex ~ body_mass_g + (1|species),
data = penguins,
family = "bernoulli",
file = "tidybayes-ggdist-3-0-m2.rds"
)The traditional way of pulling out parameters from a model in tidybayes has
been to pull them out into a long-format data frame of draws. For example, we
could use tidybayes::spread_draws() to pull draws from the global intercept
and the species-specific random offsets as follows:
m_penguin %>%
spread_draws(r_species[species,coef], Intercept)## # A tibble: 12,000 x 7
## # Groups: species, coef [3]
## species coef r_species .chain .iteration .draw Intercept
## <chr> <chr> <dbl> <int> <int> <int> <dbl>
## 1 Adelie Intercept 2.43 1 1 1 0.882
## 2 Adelie Intercept 2.57 1 2 2 1.05
## 3 Adelie Intercept 2.68 1 3 3 1.07
## 4 Adelie Intercept 3.10 1 4 4 1.34
## 5 Adelie Intercept 5.27 1 5 5 -0.678
## 6 Adelie Intercept 4.43 1 6 6 -0.451
## 7 Adelie Intercept 1.90 1 7 7 1.84
## 8 Adelie Intercept 4.93 1 8 8 -1.57
## 9 Adelie Intercept 6.06 1 9 9 -1.68
## 10 Adelie Intercept 5.68 1 10 10 -1.73
## # ... with 11,990 more rowsThis results in a 12,000-row data frame that is (1) very useful for summarizing and plotting but (2) a bit hard to read.
Using the new posterior::rvar datatype and the tidybayes::spread_rvars()
function, we can instead pull out these coefficients into long-format data frames of rvars:
m_penguin %>%
spread_rvars(r_species[species,coef], Intercept)## # A tibble: 3 x 4
## species coef r_species Intercept
## <chr> <chr> <rvar> <rvar>
## 1 Adelie Intercept 3.6 ± 2.0 0.22 ± 2
## 2 Chinstrap Intercept 3.3 ± 2.0 0.22 ± 2
## 3 Gentoo Intercept -6.4 ± 2.1 0.22 ± 2The rvar datatype acts like a normal vector or array (including supporting
many common functions and operators), but internally stores
the full sample corresponding to each cell in the array. When printed, these
draws are summarized to their mean and standard deviation. You can
plot these objects using the stat_dist_... family in ggdist:
m_penguin %>%
spread_rvars(r_species[species,coef], b_Intercept) %>%
mutate(species_intercept = b_Intercept + r_species) %>%
ggplot(aes(y = species, dist = species_intercept)) +
stat_dist_halfeye() +
xlab("Species-specific intercept")
This intercept isn’t very interpretable, so let’s instead use it with the
slope (the coefficient on body_mass_g) to calculate the body mass for each
species where we’d expect a 50-50 chance of an individual being male or female.
This is just based on a re-arrangement of the equation for the log-odds of
being male:
\[ \begin{align*} \textrm{logit}(\Pr(\textrm{male})=0.5) &= \beta_\textrm{body_mass}\textrm{body_mass}+\beta_\textrm{intercept}\\ \implies \textrm{body_mass} &= \frac{-\beta_\textrm{intercept}}{\beta_\textrm{body_mass}} & \textit{because }\textrm{logit}(0.5)=0 \end{align*} \]
We can overlay this crossing-point on the orginal dotplot. We’ll also use the
fact that side can now vary as an aesthetic to plot the dotplots flipped
for male versus female penguins (described in more detail in the next section):
m_penguin %>%
spread_rvars(r_species[species,coef], b_Intercept, b_body_mass_g) %>%
mutate(species_intercept = b_Intercept + r_species) %>%
mutate(species_50pct = -(species_intercept)/b_body_mass_g) %>%
ggplot(aes(y = species)) +
stat_dots(
aes(
x = body_mass_g, color = sex,
side = ifelse(sex == "male", "top", "bottom")
),
shape = 19, size = 0, data = na.omit(penguins), scale = 0.5
) +
stat_dist_pointinterval(aes(dist = species_50pct)) +
scale_color_brewer(palette = "Set2", guide = guide_legend(reverse = TRUE))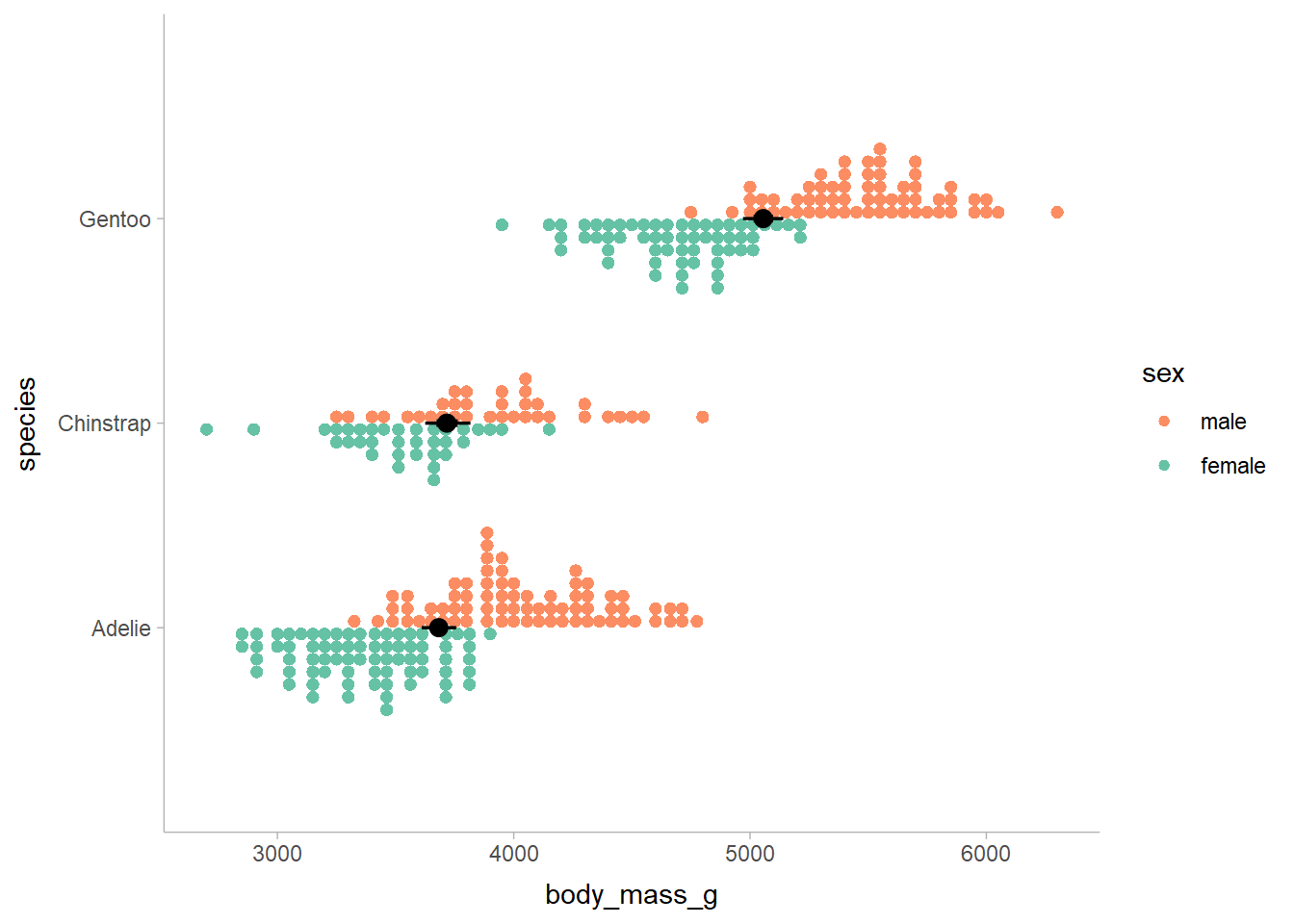
We can also use modelr::data_grid() to made a grid of predictors and then use
add_epred_rvars() to add random variables representing draws
from the expectation of the posterior predictive distribution to it. This is similar to
how add_fitted_draws() (now renamed to add_epred_draws(), see note below)
works in the classic tidybayes workflow, but instead
of returning long data frames of draws, it uses the rvar datatype to
encapsulate the draws for each row of the prediction grid. This makes the
output much easier to read (I’ll also show an example of visualizing this
in the next section):
penguins %>%
data_grid(
species,
body_mass_g = seq_range(body_mass_g, n = 50)
) %>%
add_epred_rvars(m_penguin, value = "Pr(sex = male)")## # A tibble: 150 x 3
## species body_mass_g `Pr(sex = male)`
## <fct> <dbl> <rvar>
## 1 Adelie 2700 0.0011 ± 0.00098
## 2 Adelie 2773. 0.0018 ± 0.00148
## 3 Adelie 2847. 0.0029 ± 0.00222
## 4 Adelie 2920. 0.0048 ± 0.00332
## 5 Adelie 2994. 0.0079 ± 0.00495
## 6 Adelie 3067. 0.0131 ± 0.00733
## 7 Adelie 3141. 0.0215 ± 0.01076
## 8 Adelie 3214. 0.0353 ± 0.01558
## 9 Adelie 3288. 0.0575 ± 0.02212
## 10 Adelie 3361. 0.0926 ± 0.03054
## # ... with 140 more rowsYou can also convert to/from the data-frame-of-rvars format and the long-data-frame-of-draws
format using nest_rvars() and unnest_rvars().
See the tidy-posterior
vignette for more on extracting and visualizing rvars with tidybayes.
Some of the core tidying functionality in tidybayes has also been rebuilt on top of posterior, which means tidybayes should support even more model types and benefit from efficiency improvements in posterior. This means that cmdstanr is now supported, for example.
Side, justification, and scale are now aesthetics
For geom_slabinterval(), side, justification, and scale can now be
used as aesthetics instead of parameters, allowing them to vary across slabs
within the same geom.
These changes make it possible to do things like recreate Ladislas Nalborczyk’s
logit dotplot without having
to manually figure out dotplot binwidths; here we restrict both dotplots to
less than 1/3 of the available space with scale = 1/3 and geom_dots()
does the hard work of figuring out how big the dots should be to achieve that:
penguins %>%
filter(species == "Gentoo") %>%
data_grid(
species = "Gentoo",
body_mass_g = seq_range(body_mass_g, n = 50, expand = 0.1)
) %>%
add_epred_rvars(m_penguin, value = "Pr(sex = male)") %>%
ggplot(aes(x = body_mass_g)) +
stat_dots(
aes(
y = as.numeric(sex == "male"),
side = ifelse(sex == "male", "bottom", "top")
),
data = filter(penguins, species == "Gentoo"),
na.rm = TRUE, scale = 1/3
) +
stat_dist_lineribbon(
aes(dist = `Pr(sex = male)`),
alpha = 1/4, fill = "#08306b"
) +
coord_cartesian(expand = FALSE) +
scale_fill_brewer() +
labs(
title = "stat_dots(scale = 1/3)",
subtitle = 'aes(side = ifelse(sex == "male", "bottom", "top"))',
x = "Body mass (g) of Gentoo penguins",
y = "Pr(sex = male)"
)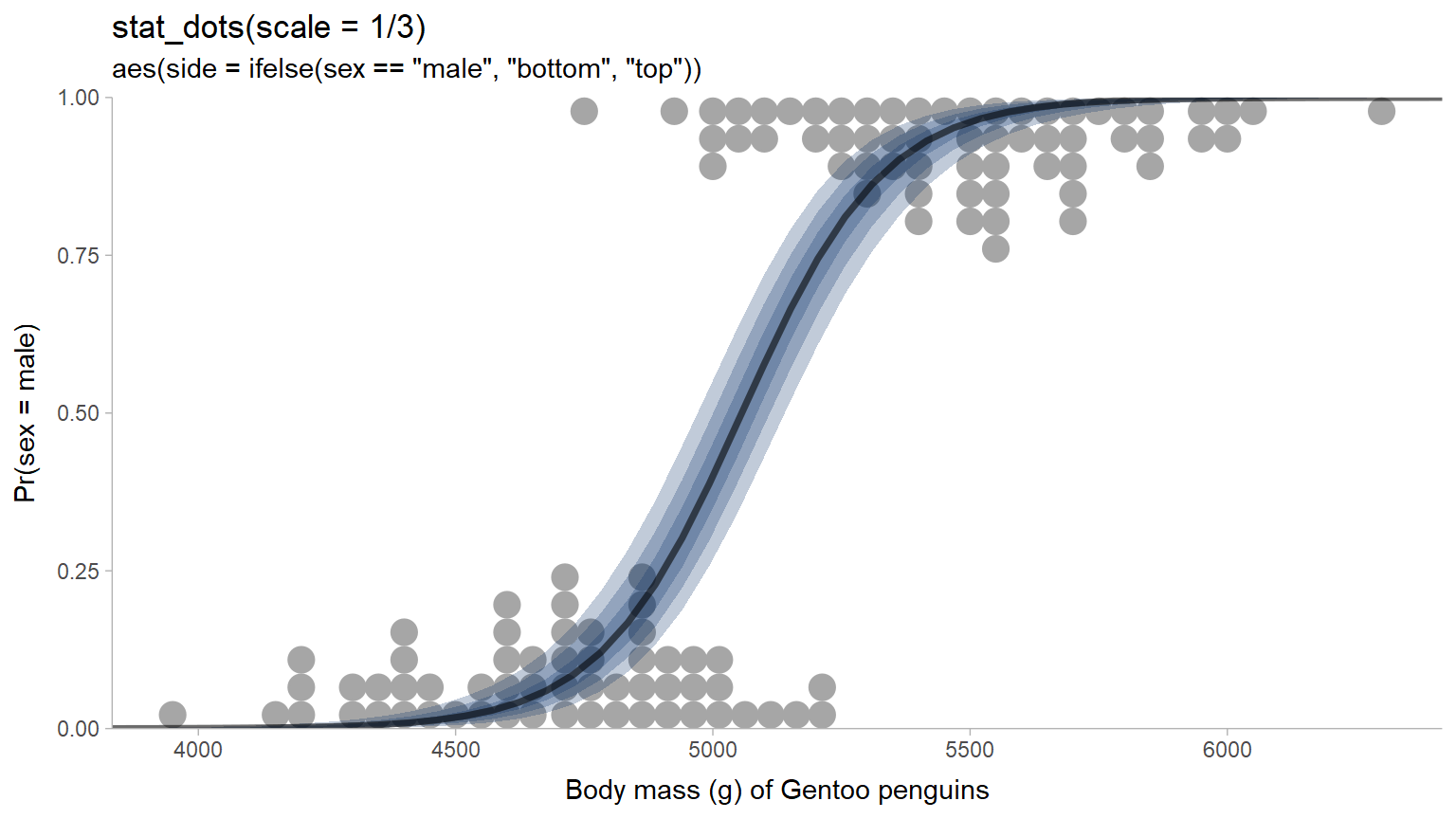
This example also shows the use of rvars with stat_dist_lineribbon().
Making scale, side, and justification into aesthetics was precipitated by
a discussion with Dominique Makowski
and Brenton Wiernik in their attempts to recreate the logit dotplot for the
see package.
Re-organization of tidybayes prediction functions
The [add_]XXX_draws() functions (predicted_draws(), add_predicted_draws(), etc)
have been substantially restructured:
add_fitted_draws()andfitted_draws()are now deprecated, along with theirscale = c("response", "linear")argument. Several years’ teaching experience has demonstrated that “fitted” is a very confusing name for students. Use the more-specific [add_]linpred_draws()if you want draws from the linear predictor or the new [add_]epred_draws()if you want draws from the expectation of the posterior predictive (which is whatfitted_draws()was most typically used for).- Arguments for renaming output columns are now all called
value, but retain function-specific default column names. E.g. thepredictionargument forpredicted_draws()is now spelledvaluebut has a default of".prediction". One breaking change is that the default output column forlinpred_draws()is now".linpred"instead of".value". This should make it easier to combine outputs across multiple functions while also making it easier to remember the name of the argument that changes the output column name. - The
nargument is now spelledndraws* to be more consistent with terminology in theposteriorpackage and to prevent partial argument name matching bugs withnewdata. - The first argument to all of these functions is now
objectinstead ofmodel, in order to match with argument names inposterior_predict(), etc. This was necessary to prevent partial argument name matching bugs with certain model types inrstanarmthat have anmargument to their prediction functions.
See the function documentation or examples from the vignettes for more.
Along with this restructuring, the epred_draws(), linpred_draws(), and
predicted_draws() functions should now support any models that implement
posterior_epred(), posterior_linpred(), and posterior_predict() so long
as they take a newdata argument.
New dodging behavior for geom_slabinterval
The positioning of geom_slabinterval() family geoms is now a bit different,
which should make it easier to line up intervals with other geoms when using
position_dodge(). Here’s an example of using position = "dodge" with
ggdist::stat_dist_halfeye(), ggplot2::geom_rect(), and ggplot2::geom_point():
dist_df %>%
ggplot(aes(
x = "a", dist = dist_normal(mean, sd), fill = group, color = group
)) +
stat_dist_halfeye(position = "dodge", color = "black") +
geom_rect(
aes(xmin = 1, xmax = 2, ymin = -3, ymax = 7),
position = "dodge",
alpha = 0.1
) +
geom_point(
aes(y = 0),
position = position_dodge(width = 1),
shape = 1, size = 4, stroke = 1.5
) +
scale_fill_brewer(palette = "Set2") +
scale_color_brewer(palette = "Dark2")
Notice how the points from geom_point() line up with the horizontal positions
of the intervals: in previous versions of ggdist this would not work.
However, the slabs do not perfectly line up with the dodged bounding boxes
shown with geom_rect(). This is because ggplot2::position_dodge() does
not preserve the relative positions of points and bounding boxes when dodging.
This can cause slabs to get close to (or in extreme cases, be drawn slightly
outside) plot bounds.
If you encounter this problem, the new ggdist::position_dodgejust(),
a “justification-preserving dodge”, can be used:
dist_df %>%
ggplot(aes(
x = "a", dist = dist_normal(mean, sd), fill = group, color = group
)) +
stat_dist_halfeye(position = "dodgejust", color = "black") +
geom_rect(
aes(xmin = 1, xmax = 2, ymin = -3, ymax = 7),
position = "dodge",
alpha = 0.1
) +
geom_point(
aes(y = 0),
# use position_dodgejust here to match the justification
# of the point to the halfeye when dodging
position = position_dodgejust(width = 1, justification = 0),
shape = 1, size = 4, stroke = 1.5
) +
scale_fill_brewer(palette = "Set2") +
scale_color_brewer(palette = "Dark2")
These changes to dodging were precipitated by some feedback from Brenton Wiernik.
Improvements for package developers
Two big changes in ggdist are aimed at making it easier for other package developers to build on it:
The automatic bin selection algorithm and and binning algorithm used by
geom_dotsinterval()have been factored out and exported asggdist::find_dotplot_binwidth()andggdist::bin_dots().Dependencies have been substantially reduced in both ggdist and tidybayes. Particularly in ggdist the hope is that the geoms will now be more suitable for use by other package developers that want to maintain smaller dependency trees (thanks to Brenton Wiernik for the help).
Other new features and changes
Other new features include:
Substantial improvements to the documentation of aesthetics and computed variables in
geom_slabinterval(),stat_slabinterval(), and company, listing all custom aesthetics, computed variables, and their usage. See the aesthetics documentation or the computed variables documentation.Previously,
curve_interval()used a common (but naive) approach to finding a cutoff on data depth to identify the X% “deepest” curves, simply taking the envelope around the X% quantile of curves ranked by depth. This is quite conservative and tends to create intervals that are too wide;curve_interval()now searches for a cutoff in data depth such that X% of curves are contained within its envelope (#67).
For more information on other minor changes, see the ggdist changelog or the tidybayes changelog.